1 unstable release
| 0.0.1 | Oct 21, 2020 |
|---|
#33 in #biology
285KB
7.5K
SLoC
protein
Protein structural Biology in Rust.
NOTE: This crate is in early development and the API has not yet been stabilized, so do not use this crate in production. If you have any suggestions, please don't hesitate to open an issue or make a PR!
Components
The protein crate essentially re-exports the components listed below. Depending on the use case, you may want to only use some of them.
| crate | Links |
|---|---|
| protein-core | |
| protein-get | |
| protein-io | |
| protein-analysis |
Example
Let's read a protein structure from a PDB file and draw a Ramachandran plot!
use csv::Writer; // the crate `csv` is required if you want to output csv
use protein::{
analysis::model::ModelAnalysis, // `Structure` alone only stores data.
get::get_pdb,
// Functions for analysing the `Structure` are provided by separate traits
io::parse_pdb, // the PDB parser that parses PDB file into a `Structure`
};
fn main() {
let pdbfile = get_pdb("4f7i").unwrap();
let structure = parse_pdb(&pdbfile).unwrap();
let (phis, psis) = structure.models[0].ramachandran();
// the `.ramachandran()` function is provided by the `ModelAnalysis` trait
// this produces vectors of phi and psi angles in radians
// the code below is used to output csv, which is optional
let mut wtr = Writer::from_path("examples/ramachandran.csv").unwrap();
wtr.write_record(&["phi", "psi"]).unwrap();
for (&phi, &psi) in phis.iter().zip(psis.iter()) {
wtr.write_record(&[phi.to_string(), psi.to_string()])
.unwrap()
}
wtr.flush().unwrap();
}
This will produce a csv file containing two columns representing phi and psi angles. Then we can read the csv file in R and plot it (unfortunately I am not familiar with any graphing libraries in Rust):
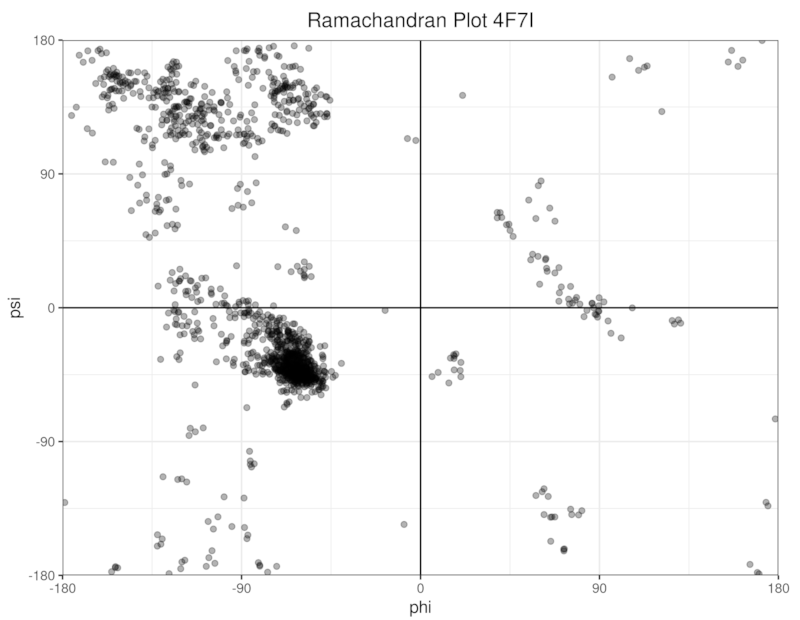
You can directly run the above example using cargo run:
cargo run --example ramachandran
Dependencies
~2–3MB
~56K SLoC