5 releases
Uses new Rust 2024
| 0.2.3 | Mar 30, 2025 |
|---|---|
| 0.2.2 | Mar 30, 2025 |
| 0.2.1 | Mar 30, 2025 |
| 0.2.0 | Mar 30, 2025 |
| 0.1.0 | Aug 28, 2023 |
#137 in Math
493 downloads per month
205KB
1.5K
SLoC
rusty_neat
Library for handling Neuroevolution of augmenting topologies.
It may not be exact implementation of that alghoritm, but it's simple, fast, and easy to use.
Innovation numers aren't generation-specific, I'm using a global look-up hashmap, so if two connections "look" the same, they have also the same number. I'm not a big fan of approach from original paper, because in my test cases innovations were exploding. Moreover, continous variant is borderline impossible to achieve without at least some modifications.
Network visual representation
Image generation is behind "visu" feature, type is derived from path (svg, png, jpg).
Blue connections are recurrent, thickness represents weight, and node 0 is a bias. Gating visualisation is something that I want to add, but I'm open to suggestions how to display it.
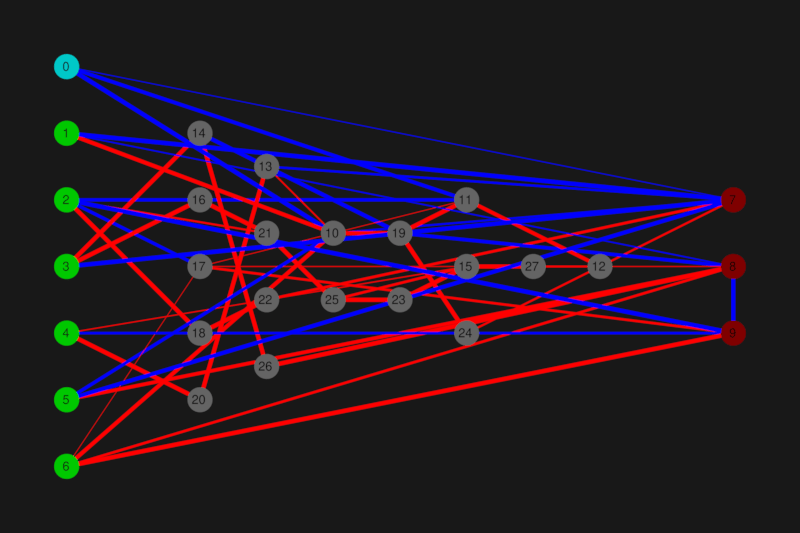
What it does?
In short, you start by creating input and output nodes, without any connections, etc. Then, through randomly evolving, there are added intermediate nodes and connections, which characteristics are randomly changed.
Features
- Evolution based on speciation and crossovers.
- Each connection has an optional gating node.
- Two handlers, one for generation-based enviroments, and second for more continous work, eg networks generate offspring on-the-run.
- Pruning (swichable): each mutation instead of expanding network, removes either node or connection (ratio is settable) in a non-destructive manner.
- Expandable I/O topology. It allows to train network on simplest possible set of inputs, and then gradually expand it's abilities.
- Network is divided in layers based on feedforward connections, which allows for quicker stabilisation time than in original neat. Solely recurrent nodes are placed in the same layer as closest "normal" node. It is kinda arbitrary, but due to chaotic neat nature (there is no cycle-prevention) it's impossible to work-out perfect calculation order.
- Network post-processing, used on evolved network to simplify it (in-progress)
How to use it?
Create large amount of agents, each with each own neural networks. Let simulation run ( or whatever you are using it for ) and after set amount of time choose best of them to be parents of next generation (intermittent mode). Alternatively, you can make agents spawn children after, for example, surviving and getting set amount of points, which nature depends on use case (continous mode).
Next generation should be created by crossing two parents, depending on the mode it looks slightly different, but at least one parent is choosed based on fitness probabillity distribution inside of species.
Inserting saved network into ongoing neat is something that I'm working on. At the moment it isn't possible due to different innovation numbers.
If docs aren't enough, or you have any feature request, feel free to reach out directly to me.
Infinite length evolution
By gradually including harder to utilise inputs networks can learn complex enviroments with relatively small agents amount. One possible downside to that approach is long training time, which could result in excessive network size. However thanks to pruning ability, size can be kept at minimum at all times, no matter training length.
Here you have example project that uses it to train "cars" ride along random track: https://github.com/HVisMyLife/neat_race.
There is also ecosystem simulation that uses continous variant, but it's currently deep in development hell: https://github.com/HVisMyLife/sectarii
Exaple usage:
use std::fs::File;
use std::io::Write;
use rusty_neat::{NN, ActFunc, visu};
use rusty_neat::NeatIntermittent;
fn main() {
let gens = 100;
let size = 10;
let mut nn = NN::new(2, 2, Some((1,1)), true, 0.75, ActFunc::HyperbolicTangent,
&[ActFunc::HyperbolicTangent, ActFunc::SELU, ActFunc::Sigmoid] );
nn.set_chances(&[0, 20, 5, 10, 3, 0, 0, 0]);
let mut handler = NeatIntermittent::new(&nn, size, 5);
handler.species_amount = 2;
handler.agents.iter_mut().enumerate().for_each(|(_,a)| a.fitness = 100. );
handler.forward(&vec![vec![1.;2]; size]);
handler.add_input();
handler.add_output(&ActFunc::HyperbolicTangent);
for i in 0..gens {
handler.speciate();
handler.next_gen();
handler.mutate(None);
handler.forward(&vec![vec![1.;3]; handler.agents.len()]);
println!("g:{}", i)
}
handler.agents.iter().enumerate().for_each(|(i, a)| {
visu(a, Some(&format!("{}.svg", i)));
a.save(&format!("{}.toml", i));
let mut file = File::create(
&("nn".to_string() + &i.to_string() + ".toml")).unwrap();
file.write_all(format!("{:?}", a).as_bytes()).unwrap();
} );
handler.set_pruning(true, 0.33);
println!("M");
for i in 0..gens {
handler.speciate();
handler.mutate(None);
handler.forward(&vec![vec![1.;8]; handler.agents.len()]);
println!("r:{}", i)
}
handler.agents.iter().enumerate().for_each(|(i, a)| {
visu(a, Some(&format!("{}.svg", i)));
visu(a, Some(&format!("{}.png", i)));
let mut file = File::create(
&("nn".to_string() + &i.to_string() + "_pruned.toml")).unwrap();
file.write_all(format!("{:?}", a).as_bytes()).unwrap();
} );
}
Possible mutations it's order, and default chances:
200 => modify one of connections weight,
20 => add new random connection,
5 => add new random node,
10 => add gating node to connection,
3 => romove gating node from connection,
0 => connection_enable,
0 => connection_disable,
0 => change one of nodes activation function,
Struct NN supports serialization and deserialization through serde.
Dependencies
~4.5–6MB
~116K SLoC