1 unstable release
| 0.1.0 | May 20, 2024 |
|---|
#12 in #variance
17KB
349 lines
molcv
This repository contains a Python package and a Rust crate for calculating the circular variance of protein residues.
The following is an example output for the 2H1L structure, where red corresponds to a circular variance of 1 and dark blue to 0.
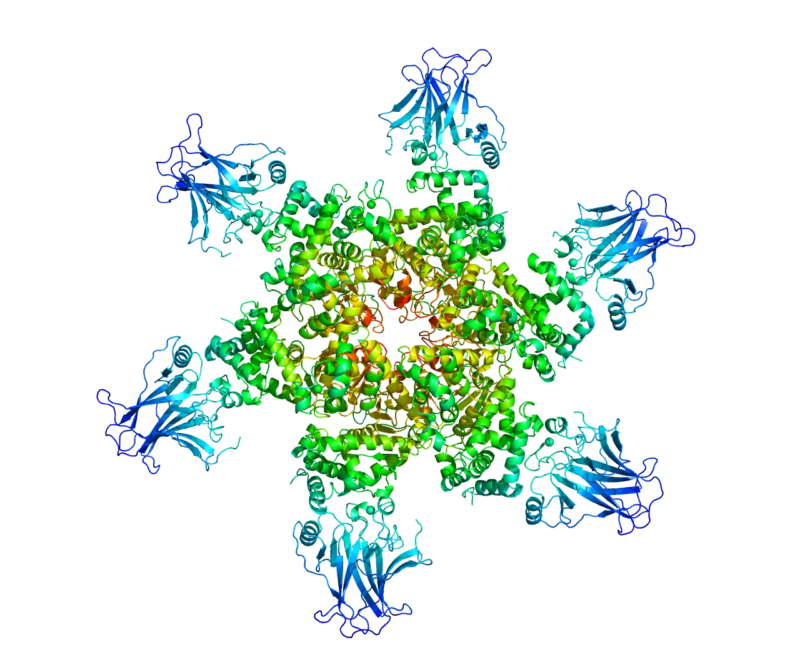
The Rust crate uses WebGPU to perform calculations on the GPU while providing support for all major platforms. The Python package contains bindings to the Rust crate.
Usage
Installation
$ cargo add molcv
# Or
$ pip install molcv
Usage
# CLI
$ molcv input.pdb --pdb-output-path output.pdb --cutoff 100
$ molcv input.pdb --npy-output-path output.npy --cutoff 50 --cutoff 100
Dependencies
~11–42MB
~650K SLoC